August 2, 2019 | Jola Glotzer
Cataloguing Drosophila matrisome
Fresh from the e-press: CBC Awardees, Sally Horne-Badovinac (UChicago) and Alexandra Naba (UIC) propose a structural and functional classification of the genes encoding Drosophila extracellular matrix (ECM) and ECM-associated proteins
A fruit fly — Drosophila melanogaster — has been used as a genetic model system to study a multitude of the developmental and physiological processes including the mechanisms underlying ECM protein secretion, ECM assembly, and ECM roles in the organism’s biology.
In a paper published today in bioRxiv, Sally Horne-Badovinac (UChicago) and Alexandra Naba (UIC), catalogue 641 ECM-related Drosophila genes according to the structural and functional properties of the ECM proteins they encode. The list has been made available to the entire scientific community as an open source resource and it is intended to help unify studies of different biological processes in a systematic way.
Horne-Badovinac, second senior author on the publication, is Associate Professor in the Department of Molecular Genetics and Cell Biology at UChicago. Together with a postdoc in her lab, Allison Zajac, she is a recipient of a CBC Postdoctoral Research Award (2016). Naba, senior and corresponding author on the paper is Assistant Professor in the Department of Physiology and Biophysics, UIC, and Associate Member of the UIC Cancer Center. She is a recipient of a recent CBC Catalyst Award (2018) for the project, “Quantitative Matrisomics: A Novel Approach to Decipher Fibrosis.” The Catalyst Award is acknowledged to have partially funded the current study. Congratulation to all authors on the publication!
Publication attributed to the CBC funding*:
Davis MN, Horne-Badovinac S, Naba A. In-silico definition of the Drosophila melanogaster matrisome. bioRxiv preprint first posted online Aug. 2, 2019. (www.biorxiv.org)
ABSTRACT:
The extracellular matrix (ECM) is an assembly of hundreds of proteins that structurally supports the cells it surrounds and biochemically regulates their functions. Drosophila has emerged as a powerful model organism to study fundamental mechanisms underlying ECM protein secretion, ECM assembly, and ECM roles in pathophysiological processes. However, as of today, we do not possess a well-defined list of the components forming the ECM of this organism. We previously reported the development of computational pipelines to define the matrisome – the ensemble of genes encoding ECM and ECM-associated proteins – of humans, mice, zebrafish and C. elegans. Using a similar approach, we report here that the Drosophila matrisome is composed of 641 genes. We further classify these genes into different structural and functional categories, including an expanded way to classify genes encoding proteins forming apical ECMs. We illustrate how having a comprehensive list of Drosophila matrisome proteins can be used to annotate large proteomic datasets and identify unsuspected roles for the ECM in pathophysiological processes. Last, to aid the dissemination and usage of the proposed definition and categorization of the Drosophila matrisome by the scientific community, our list has been made available through three public portals: The Matrisome Project, FlyBase, and GLAD.
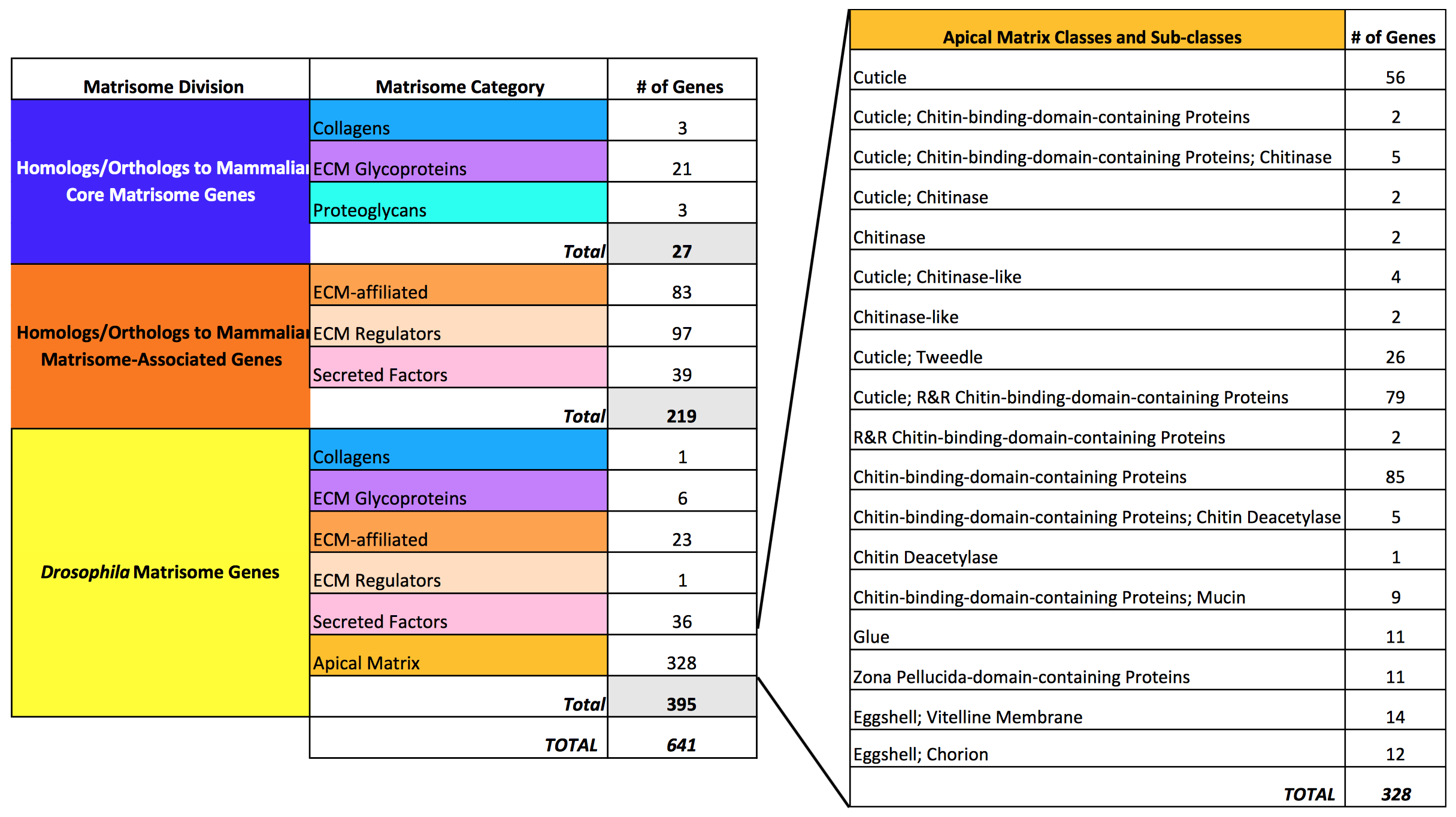
The Drosophila matrisome. (Source: www.biorxiv.org)
ACKNOWLEDGMENTS:
This work was supported by a start-up fund from the Department of Physiology and Biophysics at UIC and a Catalyst Award from the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust to AN, and by grants from the American Cancer Society (RSG-14-176) and National Institutes of Health (R01-GM126047) to SHB.
Featured CBC Community member(s):
Sally Horne-Badovinac (UChicago)
- CBC Postdoctoral Research Award (2016):
▸ Structural Reorganization of the ER During Type IV Collagen (Col IV) Folding
PIs: Allison Zajac (postdoc) and Sally Horne-Badovinac (UChicago) - CBC Scholars “Loop Connections” Seminar (2012):
▸ The Misshapen kinase negatively regulates integrin levels to promote collective cell migration in Drosophila
Sally Horne-Badovinac (UChicago) — Invited Speaker
Alexandra Naba (UIC)
- *CBC Catalyst Award (2018):
▸ Quantitative Matrisomics: A Novel Approach to Decipher Fibrosis
PIs: John Varga (NU) and Alexandra Naba (UIC)
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
April 5, 2019
▸ Keep in line, epithelium!
Deciphering the mechanisms of epithelial migration – CBC Community member Sally Horne-Badovinic, UChicago, publishes in Current Biology

