November 4, 2013
A Genetic Map for Complex Diseases
In 2012, Andrey Rzhetsky’s team received a CBC Lever Award to help fund the Conte Center for Computational Neuropsychiatric Genomics. A year later, on September 26, 2013, the team reported in Cell the development of a “genetic map” of associations between Mendelian disease loci and co-occurrence of “complex diseases” such as autism, schizophrenia, diabetes and heart disease. The “map” promises to become useful in genetic and clinical evaluations of the risk of developing complex diseases and in understanding therapeutic efforts to cope with such illnesses.
The study of the human genome was supposed to uncover a “gene for everything.” As long as we looked hard enough, scientists could find gene mutations that caused any disease and figure out a way to fix them. Easy, right? Unfortunately most diseases aren’t caused by single gene mutations. Although heavily studied, the specific genetic causes of “complex diseases,” a category of disorders which includes autism, diabetes and heart disease, are still largely unknown due to byzantine genetic and environmental interactions.
Now, scientists from the University of Chicago have created one of the most expansive analyses to date of the genetic factors at play in complex diseases — by using disorders with known genetic causes to guide them. Analyzing more than 120 million patient records and identifying trends of co-occurrence among hundreds of diseases, they created a unique genetic map that has the potential to guide researchers and clinicians in diagnosing, identifying risk factors for and someday developing therapies against complex diseases. The work, titled “A Nondegenerate Code of Deleterious Variants in Mendelian Loci Contributes to Complex Disease Risk”, was published September 26, 2013 in Cell.
 “For the first time we’ve found that almost every complex disease has a unique set of associations with single-gene diseases. This essentially gives us ‘barcodes’ of specific gene loci, which we can use to help untangle the complex genetics of complex diseases,” said Andrey Rzhetsky, PhD (right), professor of genetic medicine and human genetics at the University of Chicago, who led the study.
“For the first time we’ve found that almost every complex disease has a unique set of associations with single-gene diseases. This essentially gives us ‘barcodes’ of specific gene loci, which we can use to help untangle the complex genetics of complex diseases,” said Andrey Rzhetsky, PhD (right), professor of genetic medicine and human genetics at the University of Chicago, who led the study.
The majority of human diseases are complex and caused by a combination of genetic, environmental and lifestyle factors. On the other end of the spectrum are Mendelian diseases such as cystic fibrosis and sickle-cell anemia, which are caused by abnormalities to a single gene. Some Mendelian disorders are known to predispose patients to certain complex diseases, but these co-occurrences have thus far only been studied on a small-scale basis.
To expose any underlying shared genetic structures between these disease categories, Rzhetsky and his team developed computational algorithms to parse more than 120 million patient billing records from hospitals systems across the U.S. and from nearly the entire population of Demark. They looked for trends in comorbidity, or the occurrence of both complex and Mendelian disease in the same patient, that were higher than expected from random chance. They studied these correlations in 65 complex diseases affecting almost every system in the body, including arthritis, depression and lung cancer, and in 95 Mendelian disease groups (representing 213 disorders).
The team uncovered 2,909 statistically significant associations, as well as corresponding levels of relative risk between every disease pair. Some comorbidities were well known, such as the strong link between lipoprotein deficiencies and heart attack, but the vast majority were previously unknown. For example, Marfan syndrome, a connective tissue disorder, was found to have significant comorbidities with neuropsychiatric diseases such as autism, bipolar disorder and depression. Fragile X syndrome, an intellectual disability disorder, has significant associations with asthma, psoriasis and viral infection, highlighting a potential immune system dysfunction in these patients.
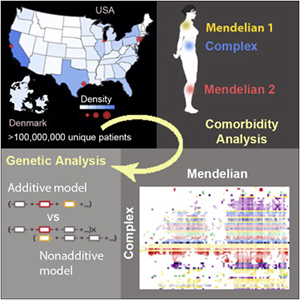 “Since the Mendelian diseases are associated with known genetic loci, we have essentially created a genetic map for complex disease using Mendelian disorders as markers,” said David Blair, a graduate student at the University of Chicago and first author on the study. “These loci represent great candidates for uncovering subtle genetic variations, some which might not directly cause Mendelian disease but still impact the risk for developing complex diseases.”
“Since the Mendelian diseases are associated with known genetic loci, we have essentially created a genetic map for complex disease using Mendelian disorders as markers,” said David Blair, a graduate student at the University of Chicago and first author on the study. “These loci represent great candidates for uncovering subtle genetic variations, some which might not directly cause Mendelian disease but still impact the risk for developing complex diseases.”
This genetic map is immediately useful for geneticists and clinicians as a gauge to the level of risk of developing complex disease among their patients with Mendelian diseases. But it also gives scientists a wealth of new data and a unique approach by which to better understand and develop therapeutics against complex diseases.
The team also discovered that genetic insults underlying Mendelian diseases do not appear to independently contribute to complex diseases but likely interact in a combinatorial way to ultimately cause the disorders.
“Individuals with multiple Mendelian disease-causing genetic variants end up having a much higher risk for a complex disease than we would predict given that the variants act in isolation,” Blair said.
The team hopes to expand their study to even more diseases and larger population data and to compare their predictions against the whole genome data of a broad population.
“Our work has the potential to open many new avenues of future research for complex diseases,” Rzhetsky said.
Adapted from: A Genetic Map For Complex Diseases, by Kevin Jiang, The University of Chicago Science Life, September 27, 2013
Andrey Rzhetsky is a co-recipient of a CBC Lever Award, Silvio O. Conte Center on the Computational Systems Genomics of Psychiatric Disorders that matched the National Institute of Mental Health grant: the Silvio O. Conte Center for Basic and Translational Mental Health Research (P-50) award. For more information about the center visit the Conte Center website.
Image credits: Andrey Rzhetsky, courtesy of Conte Center website; Graphical Abstract, adapted from: Blair DR, Lyttle CS, Mortensen JM, Bearden CF, Jensen AB, Khiabanian H, Melamed R, Rabadan R, Bernstam EV, Brunak S, Jensen LJ, Nicolae D, Shah NH, Grossman RL, Cox NJ, White KP, Rzhetsky A. A nondegenerate code of deleterious variants in mendelian Loci contributes to complex disease risk. Cell. 2013 Sep 26;155(1):70-80. (PubMed)
This work was supported by grants (1P50MH094267, NHLBI MAPGen U01HL108634-01, P50GM081892-01A1, and 2T32GM007281-39) from the National Institutes of Health and by a Lever Award from the Chicago Biomedical Consortium.
