June 17, 2019 | Jola Glotzer
Copper centers revealed with top-down mass spectrometry
Three CBC Awards contribute to today’s publication in Nature Communications!
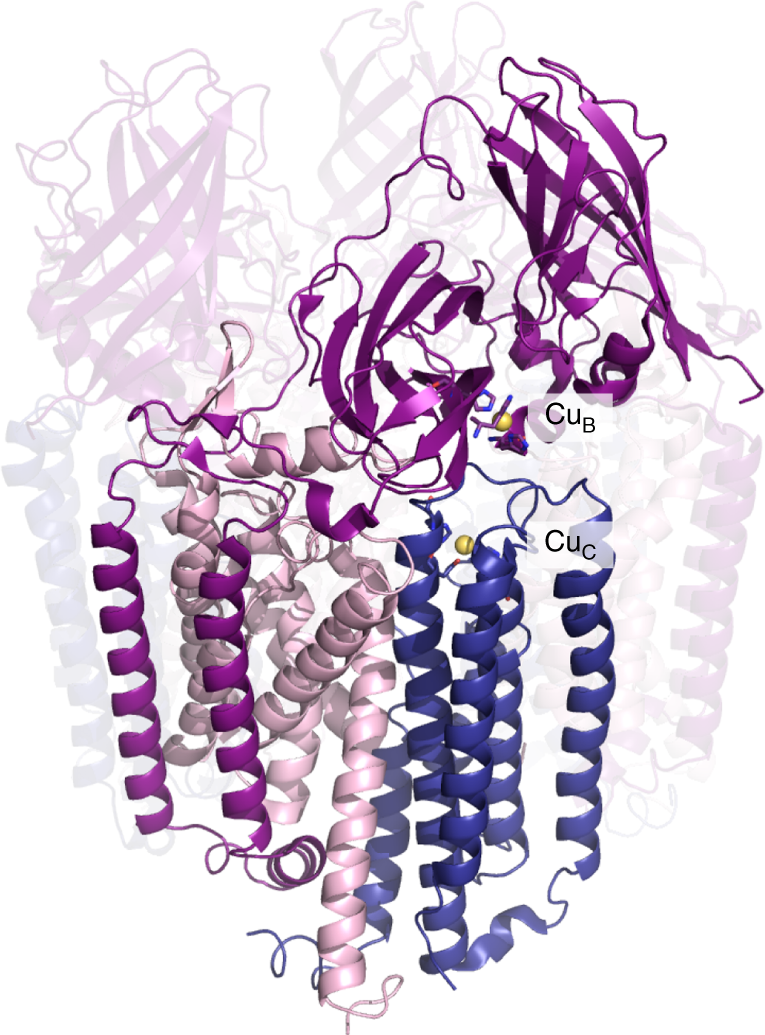
The pMMO structure and location of the metal centers. In the crystal structure of Methylocystis sp. strain Rockwell-pMMO (4PHZ), there is a monocopper center in PmoB coordinated by residues His 29, His 133, and His 135 (CuB), and a monocopper center in PmoC coordinated by Asp 129, His 133, and His 146 (CuC). PmoA, PmoB, and PmoC are shown in pink, purple, and blue, respectively, with one of the three protomers highlighted. Copper ions are shown as yellow spheres. (Source: Nature Communications.)
The CBC is pleased to share that an article published in today’s issue of Nature Communications is attributed to three CBC Awards involving four CBC awardees and/or affiliates in the study. Amy Rosenzweig, NU, last author on the paper and Rahul Purohit, the paper’s co-author and a postdoc in her lab acknowledge the first of the CBC Awards— a CBC Postdoctoral Research Award. The team received it in 2016, for the project “Cryo-electron Microscopy Studies of an Integral Membrane Copper Transporting ATPase.” The next contributor is Neil Kelleher, NU, second senior author on the publication and a CBC Senior Investigator. Kelleher was hired by NU in 2010, with help from a CBC Recruitment Resources Award. Part of the published work was also done in the NU’s Proteomics Center for Excellence (PCE), a facility directed by Kelleher and established with the CBC support. The fourth CBC affiliate who contributed to the paper is a PCE member, Paul Thomas, who helped to organize and lectured at five CBC Summer Workshops in Proteomics and Informatics (2010-2015). Finally, the authors acknowledge using the NU’s Gatan K2 direct electron detector, purchased with funds provided by the CBC in 2014 as part of the third CBC Award— an Infrastructure Initiative.
The article, titled “Native top-down mass spectrometry provides insights into the copper centers of membrane-bound methane monooxygenase,” addresses the structural and functional aspects of the particulate methane monooxygenase (pMMO), a copper-dependent, membrane metalloenzyme. pMMO oxidizes methane to methanol in methanotrophic bacteria, and comprises three subunits, PmoB, PmoA, and PmoC (see figure). The main results reported in the paper are two-fold: (1) new insights into the copper centers and (2) how native top-down mass spectrometry can be utilized to study the structure of metalloenzymes. Congratulations to all authors involved in the study!
Publication attributed to CBC funding*:
Ro SY, Schachner LF, Koo CW, Purohit R, Remis JP, Kenney GE, Liauw BW, Thomas PM, Patrie SM, Kelleher NL, Rosenzweig AC. Native top-down mass spectrometry provides insights into the copper centers of membrane-bound methane monooxygenase. Nature Communications 10, Article number: 2675. Published: 17 June 2019. (nature.com)
ABSTRACT
Aerobic methane oxidation is catalyzed by particulate methane monooxygenase (pMMO), a copper-dependent, membrane metalloenzyme composed of subunits PmoA, PmoB, and PmoC. Characterization of the copper active site has been limited by challenges in spectroscopic analysis stemming from the presence of multiple copper binding sites, effects of detergent solubilization on activity and crystal structures, and the lack of a heterologous expression system. Here we utilize nanodiscs coupled with native top-down mass spectrometry (nTDMS) to determine the copper stoichiometry in each pMMO subunit and to detect post-translational modifications (PTMs). These results indicate the presence of a mononuclear copper center in both PmoB and PmoC. pMMO-nanodisc complexes with a higher stoichiometry of copper-bound PmoC exhibit increased activity, suggesting that the PmoC copper site plays a role in methane oxidation activity. These results provide key insights into the pMMO copper centers and demonstrate the ability of nTDMS to characterize complex membrane-bound metalloenzymes.
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grants GM118035 (A.C.R.) and 1S10OD025194–01 (N.L.K). Research in this publication is also supported by a fellowship associated with the Chemistry of Life Processes Predoctoral Training Grant T32GM105538 at Northwestern University. L.F.S. is a Gilliam Fellow of the Howard Hughes Medical Institute. The Quantitative Bio-element Imaging Center at Northwestern is supported by NASA Ames Research Center NNA06CB93G. The Proteomics Center for Excellence at Northwestern is supported by NIH grant NCI CCSG P30 CA060553. The support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Northwestern University Structural Biology Facility is acknowledged. The Gatan K2 direct electron detector was purchased with funds provided by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust. Collection of electron microscopy data was supported by Chicago Biomedical Consortium Postdoctoral Research Award PDR-097 (R. P.) with support from the Searle Funds at The Chicago Community Trust.
See also:
▸ *Infrastructure Initiative: Direct Electron for the Biological CryoEM (2014)
PIs: Alfonso Mondragon and Jay Walsh, NU
Featured CBC Community member(s):
Amy Rosenzweig, NU
- *CBC Postdoctoral Research Award (2016):
▸ Cryo-electron Microscopy Studies of an Integral Membrane Copper Transporting ATPase
PIs: Rahul Purohit (postdoc) and Amy Rosenzweig (mentor; NU)
Neil Kelleher, NU
- CBC Exploratory Workshop (2013):
▸ The CBC Exploratory Workshop on Cellular Heterogeneity
Neil Kelleher (NU) – Workshop Organizer and Speaker - CBC Science Day (2011):
Neil Kelleher (NU) – Science Day Speaker - CBC Catalyst Award (2010):
▸ Phosphoproteomic Analysis of NADPH Oxidase Activation
PIs: Neil Kelleher (NU) and Richard Ye (UIC) - *CBC Recruitment Resources Award (2010):
▸ CBC Senior Investigator
Neil Kelleher (NU) - CBC Seminar (2010):
▸ Top Down Mass Spectrometry: Has Its Decade Now Come?
Neil Kelleher (NU, UIUC then) – Seminar Speaker
Paul Thomas, NU
Related:
