August 11, 2017
Big data yields surprising connections between diseases
Andrey Rzhetsky (UChicago), Edwin Cook (UIC), and Richard Morimoto (NU) are the recipients of a CBC 2012 Lever Award, which supported the establishment of Silvio O. Conte Center on the Computational Systems Genomics of Psychiatric Disorders. Recently, a group led by a member of the CBC Lever team, Andrey Rzhetsky, published their work in Nature Genetics, “Classification of common human diseases derived from shared genetic and environmental determinants.” The study, which is partially attributed to CBC funding, describes 84 brand new genetic and environmental correlations for common human diseases. Surprisingly, a CNS disease migraine “appeared to be most genetically similar to irritable bowel syndrome and most environmentally similar to cystitis and urethritis, all of which are inflammatory diseases.”
Using health insurance claims data from more than 480,000 people in nearly 130,000 families, researchers at the University of Chicago have created a new classification of common diseases based on how often they occur among genetically-related individuals.
Researchers hope the work, published this week in Nature Genetics, will help physicians make better diagnoses and treat root causes instead of symptoms.
“Understanding genetic similarities between diseases may mean that drugs that are effective for one disease may be effective for another one,” said Andrey Rzhetsky, PhD, the Edna K. Papazian Professor of Medicine and Human Genetics at UChicago who was the paper’s senior author. “And for those diseases with a large environmental component, that means we can perhaps prevent them by changing the environment.”
The results of the study suggest that standard disease classifications–called nosologies–based on symptoms or anatomy may miss connections between diseases with the same underlying causes. For example, the new study showed that migraine, typically classified as a disease of the central nervous system, appeared to be most genetically similar to irritable bowel syndrome, an inflammatory disorder of the intestine.
Rzhetsky and a team of researchers analyzed records from Truven MarketScan, a database of de-identified patient data from more than 40 million families in the United States. They selected a subset of records based on how long parents and their children were covered under the same insurance plan within a time frame most likely to capture when children were living in the same home with their parents. They used this massive data set to estimate genetic and environmental correlations between diseases.
Next, using statistical methods developed to create evolutionary trees of organisms, the team created a disease classification based on two measures. One focused on shared genetic correlations of diseases, or how often diseases occurred among genetically-related individuals, such as parents and children. The other focused on the familial environment, or how often diseases occurred among those sharing a home but who had no or partially matching genetic backgrounds, such as spouses and siblings.
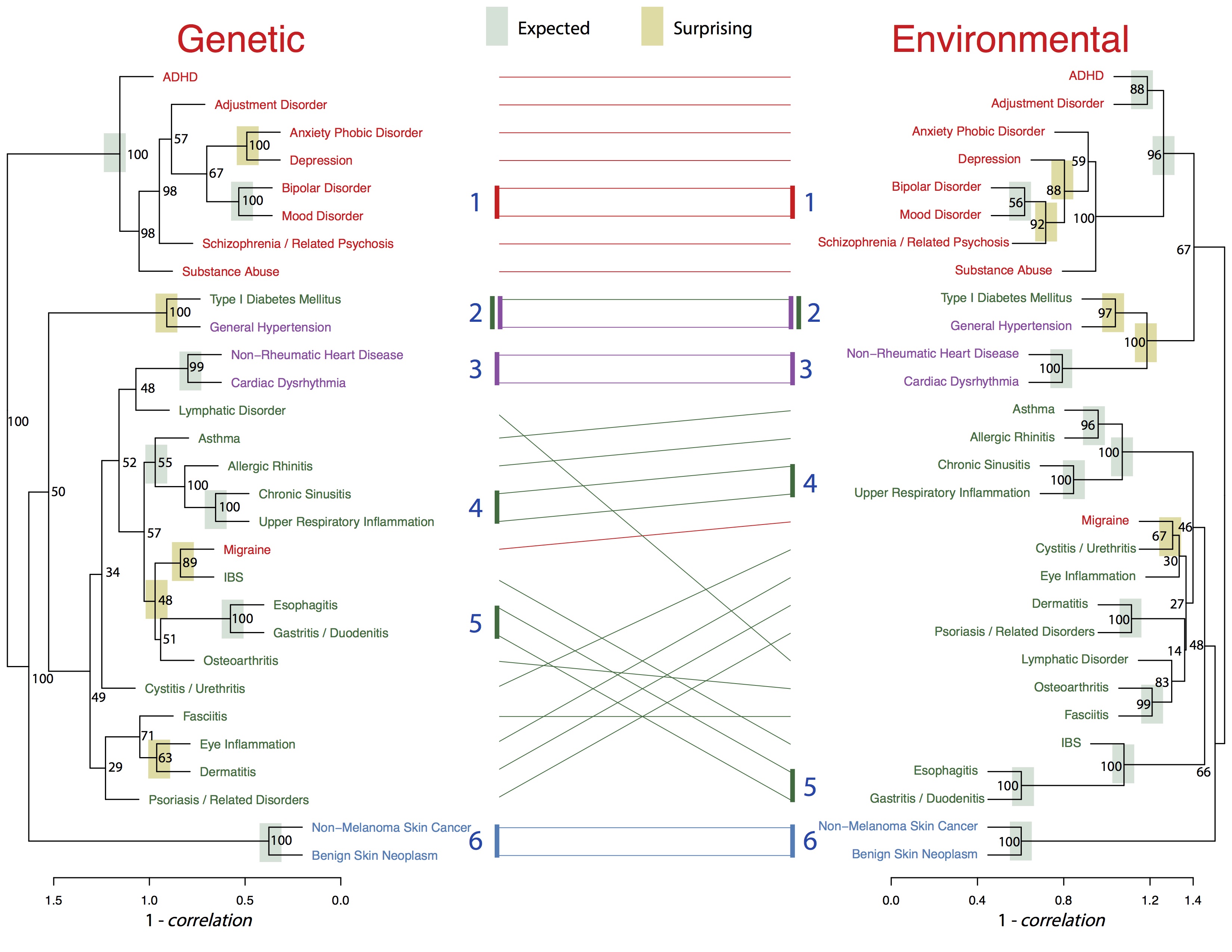
Figure: New disease classifications created by analyzing genetic and environmental correlations among family members.
The results focused on 29 diseases that were well represented in both children and parents to build new classification trees. Each “branch” of the tree is built with pairs of diseases that are highly correlated with each other, meaning they occur frequently together, either between parents and children sharing the same genes, or family members sharing the same living environment.
“The large number of families in this study allowed us to obtain precise estimates of genetic and environmental correlations, representing the common causes of multiple different diseases,” said Kanix Wang, a graduate student at UChicago and lead author of the study. “Using these shared genetic and environmental causes, we created a new system to classify diseases based on their intrinsic biology.”
Genetic similarities between diseases tended to be stronger than their corresponding environmental correlations. For the majority of neuropsychiatric diseases, such as schizophrenia, bipolar disorder and substance abuse, however, environmental correlations are nearly as strong as genetic ones. This suggests there are elements of the shared, family environment that could be changed to help prevent these disorders.
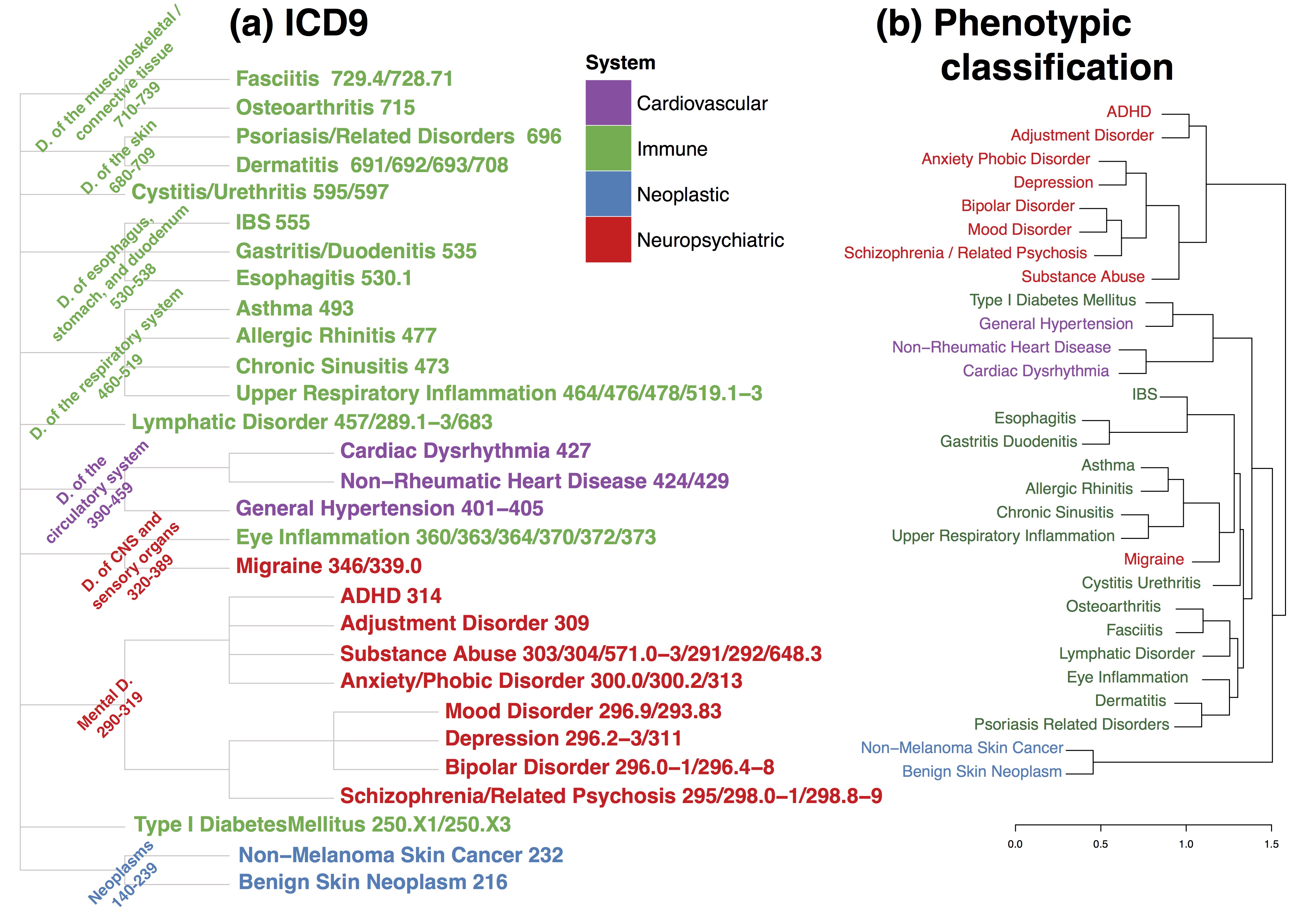
Figure: Two traditional disease classifications, ICD-9 (left) and a phenotypic model (right) based on symptoms.
The researchers also compared their results to the widely used International Classification of Diseases Version 9 (ICD-9) and found additional, unexpected groupings of diseases. For example, type 1 diabetes, an autoimmune endocrine disease, has a high genetic correlation with hypertension, a disease of the circulatory system. The researchers also saw high genetic correlations across common, apparently dissimilar diseases such as asthma, allergic rhinitis, osteoarthritis and dermatitis.
This work was funded by the DARPA Big Mechanism program under ARO contract W911NF1410333, by National Institutes of Health grants R01HL122712, 1P50MH094267*, and U01HL108634-01, and by a gift from Liz and Kent Dauten.
*NIH grant leveraged with the CBC Lever Award (2012) to Andrey Rzhetsky (UChicago), Edwin Cook (UIC), and Richard Morimoto (NU; see below).
Source: Adapted (with modifications) from the UChicago Science Life. Posted on August 7, 2017 by Matt Wood in Features.
SEE ALSO:
CBC Lever Award (2012):
PIs: Andrey Rzhetsky (UChicago), Edwin Cook (UIC), and Richard Morimoto (NU) for project:
▸ Silvio O. Conte Center on the Computational Systems Genomics of Psychiatric Disorders
Publication attributed to the CBC Lever Award (2012):
Classification of common human diseases derived from shared genetic and environmental determinants. Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. Nat Genet. 2017 Aug 7. [Epub ahead of print] (PubMed)

