March 6, 2015
Kind of Like a Hot Dog Bun
Transport molecule forms a protective structure to guide proteins to cell membrane
Congratulations to Bob Keenan and Tony Kossiakoff (UChicago) on their publication in the March 5, 2015 issue of Science: “Structure of the Get3 targeting factor in complex with its membrane protein cargo.” Kossiakoff is one of the recipients of the 2012 CBC Lever Award which helped establish the Center for Production of Affinity Reagents for Human Transcription Factors: Chicago Synthetic Antibody Pipeline (CSAP). In April 2014, Keenan received a CBC Lever-supported CSAP Pilot Project Award for the project “Antibody Phage Display of Nanodisc-embedded Membrane Proteins.”
If you take a quick look at a cell under a microscope, the membrane, a double layer of fatty lipids that separates the cell from its environment, probably doesn’t seem all that exciting. A closer inspection, however, reveals a vibrant ecosystem teeming with forests, tunnels, airports — all composed of proteins. Studded throughout a cell membrane (and also smaller membranes within the cell), these molecular machines play key roles in essential cellular functions such as signaling, catalyzing chemical reactions, and nutrient and ion transport.
In order to reside in a membrane, proteins contain hydrophobic regions that correspond to the hydrophobic lipid bilayer. But this presents a problem. During protein synthesis, which takes place inside the aqueous environment of a cell, emerging hydrophobic regions are predisposed toward clumping together or misfolding in order to avoid water. This can be very harmful, so protective cellular mechanisms immediately shield these regions as they are made and then chaperone the proteins to the membrane.
Unlike the majority of membrane proteins, tail-anchored proteins — which have roles in a wide variety of cellular functions from neurotransmitter release to insulin production — contain only one hydrophobic region that is usually the last to be synthesized. Because of this characteristic, the coordination of several molecular factors are required to prevent tail-anchored proteins from aggregating.
 The study of this pathway, known as GET (‘guided entry of tail-anchored proteins’), has been in flux. The key component of GET is a targeting factor known as Get3, which captures and shields the hydrophobic region of tail-anchored proteins. The mechanism by which Get3 performs its protective function has been debated, which leaves questions about every other step in the pathway. Studies have argued that four molecules of Get3 combine to form a tetrameric complex that encase its cargo. But other studies, including ones from Robert Keenan, PhD, (right), associate professor of biochemistry and molecular biophysics at the University of Chicago and co-author Ramanujan Hedge, PhD, of the Medical Research Council in Cambridge, have pointed to a dimeric complex involving only two subunits of Get3.
The study of this pathway, known as GET (‘guided entry of tail-anchored proteins’), has been in flux. The key component of GET is a targeting factor known as Get3, which captures and shields the hydrophobic region of tail-anchored proteins. The mechanism by which Get3 performs its protective function has been debated, which leaves questions about every other step in the pathway. Studies have argued that four molecules of Get3 combine to form a tetrameric complex that encase its cargo. But other studies, including ones from Robert Keenan, PhD, (right), associate professor of biochemistry and molecular biophysics at the University of Chicago and co-author Ramanujan Hedge, PhD, of the Medical Research Council in Cambridge, have pointed to a dimeric complex involving only two subunits of Get3.
“Our model suggests that the cell is able to shield tail-anchored proteins and get them to the right membrane at the right time through a two-subunit complex,” Keenan said. “The simple analogy is that it’s like a hot dog bun. It presents a large groove that accommodates the transmembrane region of the protein that’s being targeted.”
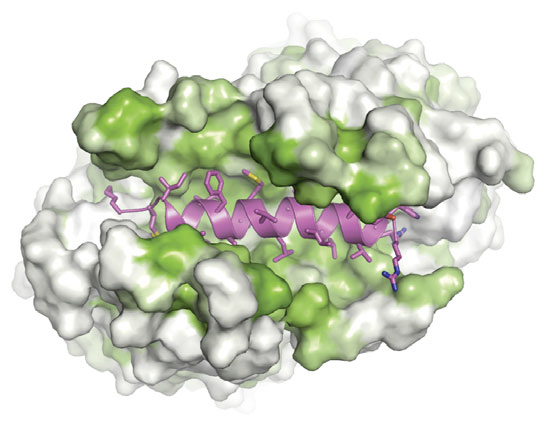
Figure: Like a hot dog and bun. A tail-anchored membrane protein (magenta) rests in a protective pocket within a Get3 complex. Green hydrophobic regions shield the substrate from the aqueous environment of a cell.
To resolve this question, a team of researchers, led by Keenan and Agnieszka Mateja, PhD, postdoctoral fellow at the University of Chicago, first assembled Get3-substrate complexes in the laboratory. Using purified components to mimic physiological conditions, they found dimeric structures. Get3 has been shown to form dimers when linked to ATP, but Get3 also naturally degrades ATP over time, which likely led to previous observations of a tetrameric Get3-substrate complexes under non-physiological conditions. When the researchers expressed a tail-anchored protein in bacteria with a modified version of Get3 that was unable to degrade ATP, only dimeric complexes were formed.
To visualize the physiologically-relevant structure, the researchers used protein crystallization to study Get3 while it was holding a hydrophobic tail-anchored protein. In collaboration with Tony Kossiakoff, PhD, professor of biochemistry and molecular biophysics at the University of Chicago, the team designed synthetic antibody fragments that bound to specific parts of the Get3 dimer. These antibodies facilitated packing of Get3 and the substrate into the crystal. Analyzing this complex atom by atom, the team found that Get3 functioned exactly as they had predicted – with two Get3 subunits protecting the tail-anchored protein in a large hydrophobic groove. They describe these findings in Science on March 5.
“There was a lot of painstaking engineering that went into this, but ultimately we defined the structure of the physiologic complex,” Keenan said. “It’s a very important biological pathway, a hard technical problem, and now only the second instance where we have a glimpse of how a hydrophobic transmembrane protein binds to one of its targeting factors. This is a fundamental insight into how a cell works.”
With the structure solved and the Get3 protective mechanism clarified, Keenan and his team are now investigating the mechanism of other steps in the GET pathway.
“The mechanistic details of the GET pathway are different if you assume a dimer model or a tetramer model,” Keenan said. “By demonstrating that the physiologically relevant targeting complex is dimeric, we get a relatively simple, elegant working model. It really clarifies thinking in this field.”
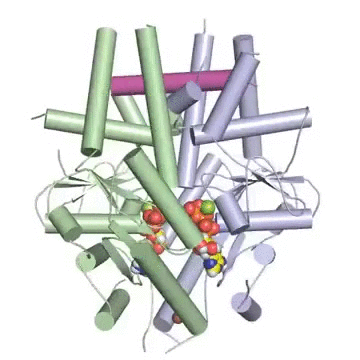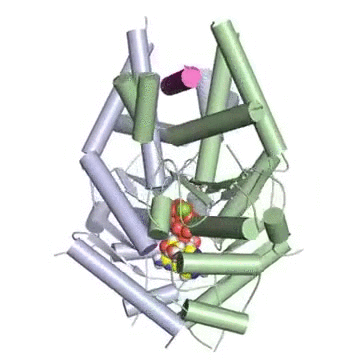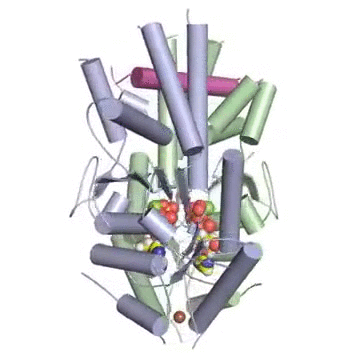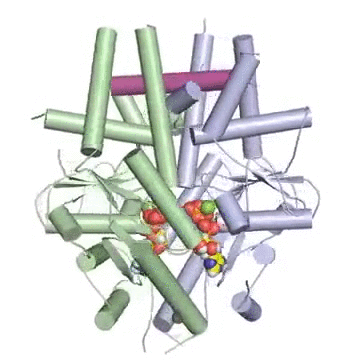
Figure: A tail-anchored membrane protein (magenta) bound to the ATP-bound (spheres) Get3 dimer (green and blue).
The study, “Structure of the Get3 targeting factor in complex with its membrane protein cargo,” was supported by the National Institutes of Health, the US Department of Energy, the UK Medical Research Council and the Chicago Biomedical Consortium.
Adapted from: "Kind of like a hot dog bun: Transport molecule forms a protective structure to guide proteins to cell membrane," Posted on March 5, 2015 by Kevin Jiang in At the Bench, Features; ScienceLife, UChicago.
Lever Award (2012):
Center for Production of Affinity Reagents for Human Transcription Factors: Chicago Synthetic Antibody Pipeline (CSAP). PIs: Anthony Kossiakoff and Geoffrey Greene (UChicago), Brian Kay (UIC), and Jason Brickner (NU)
CSAP Pilot Project (2014):
Antibody Phage Display of Nanodisc-embeded Membrane Proteins. PI: Robert Keenan (UChicago)
Publication attributed to the CSAP Pilot Project:
Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 6 March 2015 Vol. 347 no. 6226 pp. 1152-1155.
