January 14, 2015
Finding Damaged DNA — Needle in the Haystack
Congratulations to the CBC Catalyst Team: Jung-Hyun Min (UIC) and Chuan He (UChicago) on their recent publication in the January 6, 2015 issue of Nature Communications, “Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.” Min and He received the Catalyst Award in the Fall 2011 Round for a project titled “Capturing Kinetically Labile Multiprotein Assemblies on DNA by Chemical Crosslinking.”

 “The paper is the outcome of the collaboration between Prof. Chuan He and myself, catalyzed by the CBC Catalyst award,” said Min (right). “Our paper concerns the so-called “needle in the haystack” problem in genetic mechanisms, and addresses how a DNA damage recognition protein can find damaged DNA sites embedded in an enormous excess of normal DNA sites in cells.”
“The paper is the outcome of the collaboration between Prof. Chuan He and myself, catalyzed by the CBC Catalyst award,” said Min (right). “Our paper concerns the so-called “needle in the haystack” problem in genetic mechanisms, and addresses how a DNA damage recognition protein can find damaged DNA sites embedded in an enormous excess of normal DNA sites in cells.”
As summarized in the publication’s abstract: “The xeroderma pigmentosum C (XPC) complex initiates nucleotide excision repair by recognizing DNA lesions before recruiting downstream factors. How XPC detects structurally diverse lesions embedded within normal DNA is unknown. Here we present a crystal structure that captures the yeast XPC orthologue (Rad4) on a single register of undamaged DNA. The structure shows that a disulphide-tethered Rad4 flips out normal nucleotides and adopts a conformation similar to that seen with damaged DNA. 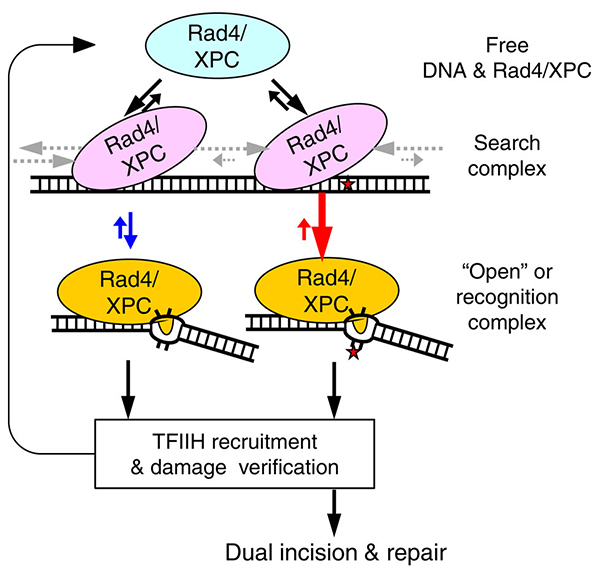 Contrary to many DNA repair enzymes that can directly reject non-target sites as structural misfits, our results suggest that Rad4/XPC uses a kinetic gating mechanism whereby lesion selectivity arises from the kinetic competition between DNA opening and the residence time of Rad4/XPC per site. This mechanism is further supported by measurements of Rad4-induced lesion-opening times using temperature-jump perturbation spectroscopy. Kinetic gating may be a general mechanism used by site-specific DNA-binding proteins to minimize time-consuming interrogations of non-target sites.”
Contrary to many DNA repair enzymes that can directly reject non-target sites as structural misfits, our results suggest that Rad4/XPC uses a kinetic gating mechanism whereby lesion selectivity arises from the kinetic competition between DNA opening and the residence time of Rad4/XPC per site. This mechanism is further supported by measurements of Rad4-induced lesion-opening times using temperature-jump perturbation spectroscopy. Kinetic gating may be a general mechanism used by site-specific DNA-binding proteins to minimize time-consuming interrogations of non-target sites.”
This schematic (left) illustrates that the lesion recognition by Rad4/XPC is controlled by the competition between Rad4’s residence time per site when it is in the ‘search’ mode (pink) and its opening time to form the ‘open’ or recognition complex (yellow). The diffusion rates that control the residence time are indicated by the grey arrows. The opening (and its reverse) rates are depicted in red and blue arrows for damaged and undamaged DNA, respectively. (Adapted from Nat Commun. 2015 Jan 6;6:5849.)
“We believe this work will not only have substantial impact on the field of genetic mechanisms, but also be of interest to general public” concluded Min.
Catalyst Award (2011): PIs: Jung-Hyun Min, PhD, UIC; Chuan He, PhD, UChicago. Capturing Kinetically Labile Multiprotein Assemblies on DNA by Chemical Crosslinking.
Publication attributed to the CBC Catalyst Award: Chen X, Velmurugu Y, Zheng G, Park B, Shim Y, Kim Y, Liu L, Van Houten B, He C, Ansari A, Min JH. Kinetic gating mechanism of DNA damage recognition by Rad4/XPC. Nat Commun. 2015 Jan 6;6:5849. (PubMed)
This work was funded by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to C.H. and J.-H.M.), NIH grants GM071440 (to C.H.) and 1R01ES019566 (to B.V.H.), NSF grants MCB-0721937 and MCB-1158217 (to A.A.), the Chancellor’s Discovery Fund (to A.A. and J.-H.M.) and a startup fund from the University of Illinois at Chicago (to J.-H.M.).
SEE ALSO:
▸ "Damaged DNA may stall patrolling molecule to initiate repair", by Jeanne Galatzer-Levy, UIC News Center, January 28, 2015
