April 18, 2014
How Immune Cells Tell Healthy and Diseased Cells Apart
Greg Tietjen, PhD, was selected as a CBC Scholar in 2012, when he was a graduate student in the Department of Biophysical Sciences & Institute for Biophysical Dynamics at the University of Chicago. He was supervised by two co-mentors, Erin Adams, associate professor of biochemistry and molecular biophysics (specializing in the immune system), and Ka Yee Lee, professor of chemistry. This unique collaboration enabled Greg to comprehensively study the role of the cell membranes lipids in the process of immunorecognition. Greg’s results were published in the March 31, 2014 issue of PNAS: “Molecular mechanism for differential recognition of membrane phosphatidylserine by the immune regulatory receptor Tim4.”
Gobbling up bad bacteria isn’t the only critical job that phagocytes, a class of amoeba-like white blood cells, have in the human immune system. They’re trash collectors too: They consume dead, dying or abnormal cells to prevent potentially harmful toxins from spreading. How these single-celled blobs recognize foreign invaders is relatively well understood, but how they identify which of the body’s own cells to destroy—essential for preventing autoimmune diseases and removing potentially cancerous cells, for example—is more opaque.
 But researchers from the University of Chicago may be closer to understanding how the body reads its own mixed signals. In a study published in the Proceedings of the National Academy of Sciences this week, they describe how they have devised new tools to uncover the inner workings of the molecular interaction at the heart of this process.
But researchers from the University of Chicago may be closer to understanding how the body reads its own mixed signals. In a study published in the Proceedings of the National Academy of Sciences this week, they describe how they have devised new tools to uncover the inner workings of the molecular interaction at the heart of this process.
One mechanism thought to be crucial to the recognition of healthy or diseased cells is the ability of phagocytes to detect a molecule known as phosphatidylserine (PS). All cells have a membrane, made of two layers of fatty acid strands known as lipids, which separates a cell from its environment. PS, a lipid itself, is a component of this membrane. In normal, healthy cells, it is sequestered into the membrane layer facing the inside of the cell. But diseased or dying cells are unable to actively move PS, and it begins to accumulate in the outside layer. Once a certain level of PS is reached, phagocytes begin to consume the cell.
Studies have shown that phagocytes use a protein receptor known as Tim4 to detect PS, but it was unknown how Tim4 finds and determines the concentration of PS in the shifting soup of lipids, proteins and other molecules that is the cell membrane. As Tim4 and PS interact at the atomic level in an ever-changing environment, identifying this mechanism seemed almost impossible.
Luckily, the unique dual-mentorship model of the UChicago Biophysics program partnered two groups perfectly suited to get at this problem. Recently graduated biophysics student Greg Tietjen brought together his two co-mentors, Erin Adams, associate professor of biochemistry and molecular biophysics (specializing in the immune system), and Ka Yee Lee, professor of chemistry.
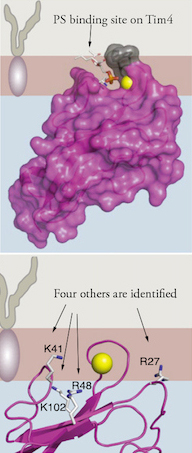
They discovered that Tim4 does not have just one site that binds to PS, but five, an arrangement which makes it sensitive to PS concentration. “We knew from crystallography data that Tim4 proteins possessed a PS binding pocket, which we thought would be like a little shark that bites onto PS,” Adams said. “It turns out that it’s the whole surface of Tim4 that engages the membrane.”
 Previous structural studies revealed a single location on the Tim4 protein that binds to PS. To test this, the team created artificial lipid bubbles—sacs made up of two layers of lipids containing a specific range of PS concentrations. They then added Tim4 proteins at carefully controlled levels. They found that Tim4 bound to PS at much higher rates than would be expected than if Tim4 bound to just one PS molecule—suggesting that each Tim4 interacted with numerous PS molecules.
Previous structural studies revealed a single location on the Tim4 protein that binds to PS. To test this, the team created artificial lipid bubbles—sacs made up of two layers of lipids containing a specific range of PS concentrations. They then added Tim4 proteins at carefully controlled levels. They found that Tim4 bound to PS at much higher rates than would be expected than if Tim4 bound to just one PS molecule—suggesting that each Tim4 interacted with numerous PS molecules.
“Although we had protein crystallography data for Tim4, it was a freeze frame, a static structure,” Lee said. “Whereas in a fluid environment such as the cell membrane, there are always fluctuations. We had to devise new tools to figure out how Tim4 engages the lipid layer.”
To further probe this interaction, especially in the hectic environment of the cell membrane, team had to devise entirely new tools. They created a single layer of lipids, containing PS, floating as a sheet in a small trough of buffered solution. Then, at the Advanced Photon Source at Argonne National Laboratory, they bounced an x-ray beam off the lipids and into a detector, which allowed them to measure electron density. When Tim4 was added to the buffered solution, the binding process affected the electron density. The researchers used these differences to calculate how the molecules were interacting.
However, this method did not provide a clear picture of what was happening at the atomic level. To gain this insight, the team turned to Benoit Roux, professor of biochemistry and molecular biophysics, and the expertise of his lab at constructing detailed atomic models via molecular dynamics simulations. Tietjen created a new suite of tools to combine these simulation models with known structural and freshly gathered x-ray data to compute exactly how Tim4 interacted with PS.
The team found that, in addition to the previously known location, four other sites on the Tim4 protein interacted with PS. While the primary site latches onto a PS molecule on the cell membrane, the other sites draw PS toward the Tim4 protein and interact in a weaker manner. This allows one Tim4 protein to detect variations in PS concentration on the membrane surface. These sites were verified by experiments that modified these locations, which had the effect of negating the ability of Tim4 to recognize more PS.
“Understanding the role of PS exposure in immune detection is important basic science, and in the context of cancers, it has tremendous significance,” Adams said. “Cancerous cells deregulate PS exposure on their outer membranes, but if we think of Tim proteins as an ‘eat-me’ signal, cancer upregulates a ‘don’t eat me’ signal.’ If we can better understand how Tims bind, we can perhaps use that to better target cancer cells and circumvent this ‘don’t eat me signal.’”
While Tim4 was the central focus of this study, other members of the Tim family of proteins also recognize PS. The team anticipates these new tools will be able to help elucidate the structures and functions of the other Tim proteins, as well as other protein-lipid interactions that have thus far eluded detailed examination.
“We plan on making our software freely accessible to the x-ray scattering community,” Lee said. “It’s one thing to have a static structure, and it’s another to be able to measure the dynamics of the situation, especially in a lipid bilayer which you can’t crystalize. This is the first paper to really detail that process using this suite of techniques, and we hope it will open the door for the next level of structural work.”
The study was supported by the National Institutes of Health, the Computation Institute and the Biological Sciences Division of the University of Chicago, Argonne National Laboratory, the National Science Foundation and the Chicago Biomedical Consortium.
Adapted from: How Immune Cells Tell Healthy And Diseased Cells Apart. Posted on March 31, 2014 by Kevin Jiang in At the Bench, ScienceLife, The University of Chicago.
CBC Scholars Award (2012):
Gregory Tietjen, Department of Biophysical Sciences & Institute for Biophysical Dynamics, The University of Chicago; Advisors: Erin Adams and Ka Yee Lee
Publication attributed to the CBC Scholars Award:
Tietjen GT, Gong Z, Chen CH, Vargas E, Crooks JE, Cao KD, Heffern CT, Henderson JM, Meron M, Lin B, Roux B, Schlossman ML, Steck TL, Lee KY, Adams EJ. Molecular mechanism for differential recognition of membrane phosphatidylserine by the immune regulatory receptor Tim4. Proc Natl Acad Sci U S A. 2014 Mar 31. (PubMed)
To learn more about the CBC Scholars Program visit the Program website.
